Extremophile organisms can withstand physicochemical stressors lethal to most others and provide models to understand life’s capacities and limitations to adapt to hostile environments. Extreme environments provide ideal systems for an integrative approach to study physiological adaptation, as their selective regimes are clearly defined and replicated, enabling hypothesis-driven tests of effects at all levels of biological organization and facilitating analysis of convergent evolution. The availability of ancestral forms living in habitats without physicochemical stressors also allows for powerful comparative approaches. Consequently, extremophiles provide unique opportunities to address major questions in contemporary biology: What mechanisms link genomic variation to fitness? Is adaptive evolution repeatable and predictable?
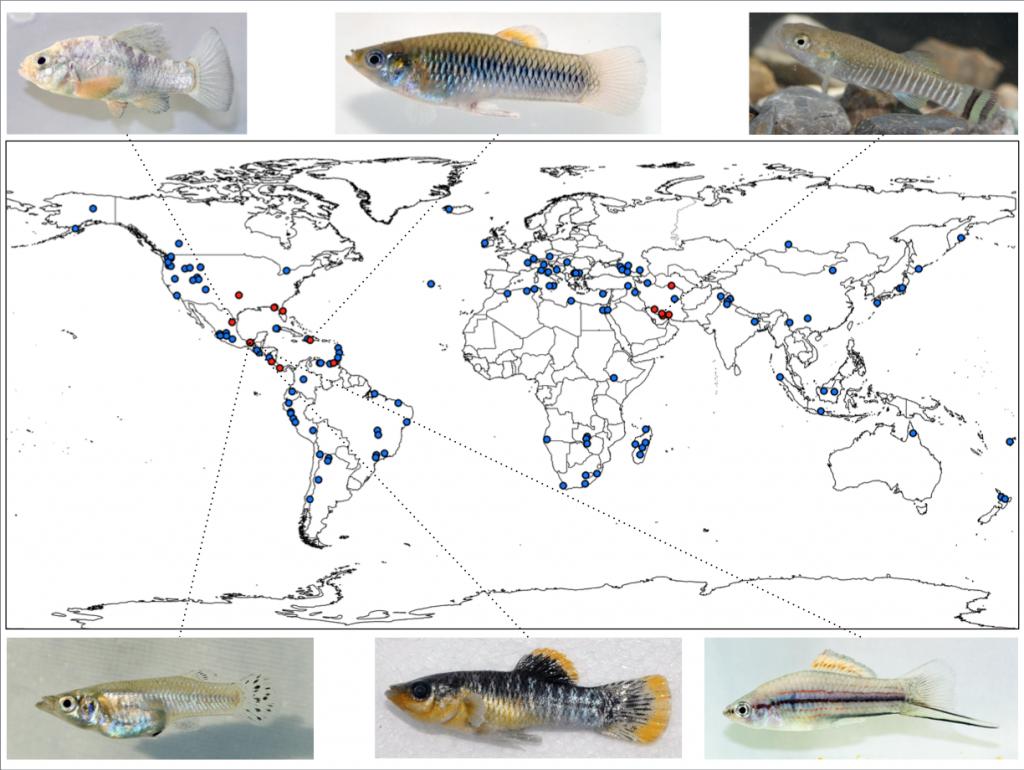
Among the most extreme freshwater ecosystems are springs rich in hydrogen sulfide (H2S), a potent respiratory toxicant lethal for most animals in micromolar concentrations. Few metazoans have successfully colonized sulfide springs, even though such springs occur wordwide. Fish of the family Poeciliidae are among the most prominent inhabitants of sulfide springs, as several species have colonized and adapted to sulfidic conditions. Most of our research investigating mechanisms underlying adaptation and speciation focuses on three evolutionarily replicated sulfide spring lineages of the genus Poecilia in Southern Mexico, while projects illuminating the nature of convergent evolution rely on a broader taxon sampling of poeciliids throughout North America, the Neotropics, and the Caribbean.
Disentangling the interactions among acclimatization, developmental plasticity, and genomic variation underlying the evolution of complex phenotypes
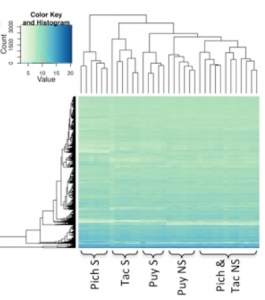 Sulfide spring fishes collected in natural populations exhibit significantly higher H2S tolerance and are characterized by complex phenotypic trait differentiation compared to ancestors in adjacent non-sulfidic populations. Phenotypic modifications include behavioral (e.g., aquatic surface respiration, aggression), physiological (e.g., metabolic rates, detoxification processes), morphological (e.g., body shape, relative organ sizes), and life history traits (e.g., offspring size and fecundity). Despite the well-characterized phenotypic differences, it remains largely unclear what mechanisms underlie trait differentiation and mediate increased sulfide tolerance. Establishing such mechanistic links is particularly challenging in ecological physiology, because environmental gradients are notoriously complex and organismal responses to physicochemical stressors occur at multiple hierarchical levels. Hence, a key research goal is to disentangle how interactions between short-term acclimatization, developmental plasticity, and genomic variation shape phenotypic differences between sulfidic and non-sulfidic populations. Current efforts combine common garden experiments, H2S exposure trials, and phenomics (including quantification of gene expression, physiological, morphological, and behavioral traits) to quantify trait co-variation and to determine what traits ultimately predict performance. In addition, we are employing population genomics approaches to identify genes and regulatory regions currently under selection and creating admixed laboratory populations that allow identifying genomic regions associated with phenotypic trait variation through quantitative genetic approaches.
Sulfide spring fishes collected in natural populations exhibit significantly higher H2S tolerance and are characterized by complex phenotypic trait differentiation compared to ancestors in adjacent non-sulfidic populations. Phenotypic modifications include behavioral (e.g., aquatic surface respiration, aggression), physiological (e.g., metabolic rates, detoxification processes), morphological (e.g., body shape, relative organ sizes), and life history traits (e.g., offspring size and fecundity). Despite the well-characterized phenotypic differences, it remains largely unclear what mechanisms underlie trait differentiation and mediate increased sulfide tolerance. Establishing such mechanistic links is particularly challenging in ecological physiology, because environmental gradients are notoriously complex and organismal responses to physicochemical stressors occur at multiple hierarchical levels. Hence, a key research goal is to disentangle how interactions between short-term acclimatization, developmental plasticity, and genomic variation shape phenotypic differences between sulfidic and non-sulfidic populations. Current efforts combine common garden experiments, H2S exposure trials, and phenomics (including quantification of gene expression, physiological, morphological, and behavioral traits) to quantify trait co-variation and to determine what traits ultimately predict performance. In addition, we are employing population genomics approaches to identify genes and regulatory regions currently under selection and creating admixed laboratory populations that allow identifying genomic regions associated with phenotypic trait variation through quantitative genetic approaches.
Understanding the physiological effects and processing of H2S: adaptation research with biomedical implications
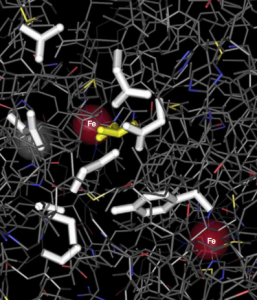 Sulfide spring fishes are not only interesting models in evolutionary biology, but they also have the potential to provide critical insights for biomedical applications. H2S has long been viewed as a toxic substance, but recent in vitro studies have documented a plethora of molecular targets and suggest that – at low concentrations – the molecule may be an important modulator of physiological processes. Accordingly, H2S is gaining increasing attention as a potential therapeutic. This has lead to a paradigm shift in H2S biology from a focus on mechanisms minimizing adverse effects of sulfide to mechanisms that maintain optimal concentrations to assure proper physiological functioning. Nevertheless, physiological processing of and organismal responses to H2S remain poorly understood. For a successful medical application, it is necessary to understand how organisms process, detoxify, and regulate sulfide, but progress toward this goal has been hampered by the availability of suitable model systems that exhibit variation in sulfide processing. We leverage sulfide spring fishes to gain an in depth understanding of organismal sulfide processing. Studying populations susceptible to H2S allows for quantifying consequences of disruptions in H2S homeostasis at all levels of biological organization (like in other systems). Adding closely related H2S-tolerant populations further allows for the detection of critical modifications in physiological processes affected by H2S. Consequently, a comparative approach provides a powerful tool to uncover physiological adaptations that are either involved in maintaining H2S homeostasis or are affected by disruptions of H2S homeostasis. We are currently combining dose-dependent exposure experiments in vivo and in vitro with physiological, biochemical, and comparative genomic analyses to uncover mechanisms involved in sulfide sequestration and oxidation as well as to investigate sulfide’s effects on the oxidative phosphorylation pathway and mitochrondrial function.
Sulfide spring fishes are not only interesting models in evolutionary biology, but they also have the potential to provide critical insights for biomedical applications. H2S has long been viewed as a toxic substance, but recent in vitro studies have documented a plethora of molecular targets and suggest that – at low concentrations – the molecule may be an important modulator of physiological processes. Accordingly, H2S is gaining increasing attention as a potential therapeutic. This has lead to a paradigm shift in H2S biology from a focus on mechanisms minimizing adverse effects of sulfide to mechanisms that maintain optimal concentrations to assure proper physiological functioning. Nevertheless, physiological processing of and organismal responses to H2S remain poorly understood. For a successful medical application, it is necessary to understand how organisms process, detoxify, and regulate sulfide, but progress toward this goal has been hampered by the availability of suitable model systems that exhibit variation in sulfide processing. We leverage sulfide spring fishes to gain an in depth understanding of organismal sulfide processing. Studying populations susceptible to H2S allows for quantifying consequences of disruptions in H2S homeostasis at all levels of biological organization (like in other systems). Adding closely related H2S-tolerant populations further allows for the detection of critical modifications in physiological processes affected by H2S. Consequently, a comparative approach provides a powerful tool to uncover physiological adaptations that are either involved in maintaining H2S homeostasis or are affected by disruptions of H2S homeostasis. We are currently combining dose-dependent exposure experiments in vivo and in vitro with physiological, biochemical, and comparative genomic analyses to uncover mechanisms involved in sulfide sequestration and oxidation as well as to investigate sulfide’s effects on the oxidative phosphorylation pathway and mitochrondrial function.
Capitalizing on a large-scale natural experiment to test hypotheses about the nature of convergent evolution
Research on replicated lineages of sulfide spring fish of the genus Poecilia has provided clear evidence for convergent evolution in physiological, morphological, and life history traits. However, the potential importance of non-convergent evolution is illustrated by molecular analyses: gene expression patterns are largely non-convergent and unique to specific sulfidic populations and H2S-tolerant cytochrome c oxidases have only evolved in a subset of replicated lineages. Consequently, both convergent and non-convergent evolution play a role in adaptation to sulfide springs, and quantitatively assessing alternative mechanisms with which different lineages have evolved to cope with high H2S concentrations is pivotal to understand evolutionary consequences of this respiratory toxicant. We are capitalizing on sulfide spring fishes that span a wide gradient of phylogenetic relationships within the family Poeciliidae to test fundamental hypotheses about the nature of convergent evolution at different levels of biological organization. Specifically, we are testing how the degree of convergence on both the phenotypic and genomic level is related to the degree of evolutionary relatedness of lineages as well as the similarity of multifarious selective regimes. We have active research projects in the US, Mexico, Costa Rica, and the Dominican Republic to investigate genomic and phenotypic differentiation in replicated population pairs of sulfide spring fishes in the genera Poecilia, Gambusia, Limia, Pseudoxiphophorus, and Xiphophorus.